|  PAINT: Leishmania Sexual Reproductive Strategies As Resolved Through Compuational Methods Designed for Aneuploid Genomes | 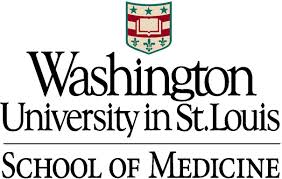 |
MAIN INDEX ANALYTICAL PIPELINE CONTACT SYSTEM REQUIREMENTS AGELESS Package Example Data | Generate A merged Allele fileThis page walks you though various steps required to generate a Merged allele file using AGELESS package.Prerequisites1) Align the fastq files to the reference genome of interest using your favourite aligner (Bowtie, BWA, Novoalign etc.)2) Sort the generated Binary alignment map (BAM) file using samtools 'sort' function 3) Generate allele files using findAlleles utility of PAINT 4) Generate list files using listAlleles utility of PAINT 5) Find parental SNVs using findSNPsSomy utility of PAINT. Extract Homozygous SNVs from the parental lines and place them in a directory. How to Run it?The findAllelesSNPpositions utility of PAINT package takes the parental homozygous SNV files and finds markers where parental lines are different from each other. It then finds the composition of all files in the listFiles directory at those markers. Type java -jar PAINT.jar findAllelesSNPpositions -h for options.
Command Examplejava -jar PAINT.jar findAllelesSNPpositions -i "../DemoData/listFiles" -j "../DemoData/snpFile/homozygous/" -o "../DemoData/otherFiles/mergedAlleles.txt" -k "vcf"OutputThe output of the findAllelesSNPpositions utility program looks as follows:
|